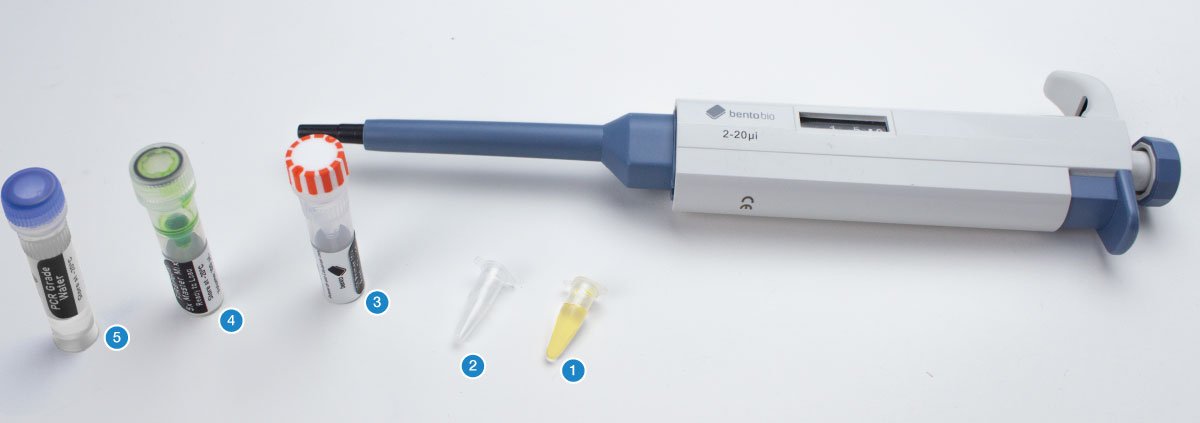
Overview
What are the possible results for this experiment?
The Cytochrome b gene is specific to different species. With the primers in this experiment we can determine the presence of the Cytochrome b gene for beef, and the Cytochrome b gene for pork.
There are two possible bands. If the sample is pure beef, we will only observe one DNA band for the bovine gene. If there is presence of pork, we will see a DNA band for the gene unique to pigs.
- Beef: the cow version of the Cytochrome b gene has been successfully copied, there is beef in the sample, and the PCR reaction has worked.
- Pork: the pig version of the Cytochrome gene has been successfully copied, which suggests there is presence of pork in the sample.