- Portable and compact: Perfect for fieldwork, pairing seamlessly with the MinION for on-site sequencing.
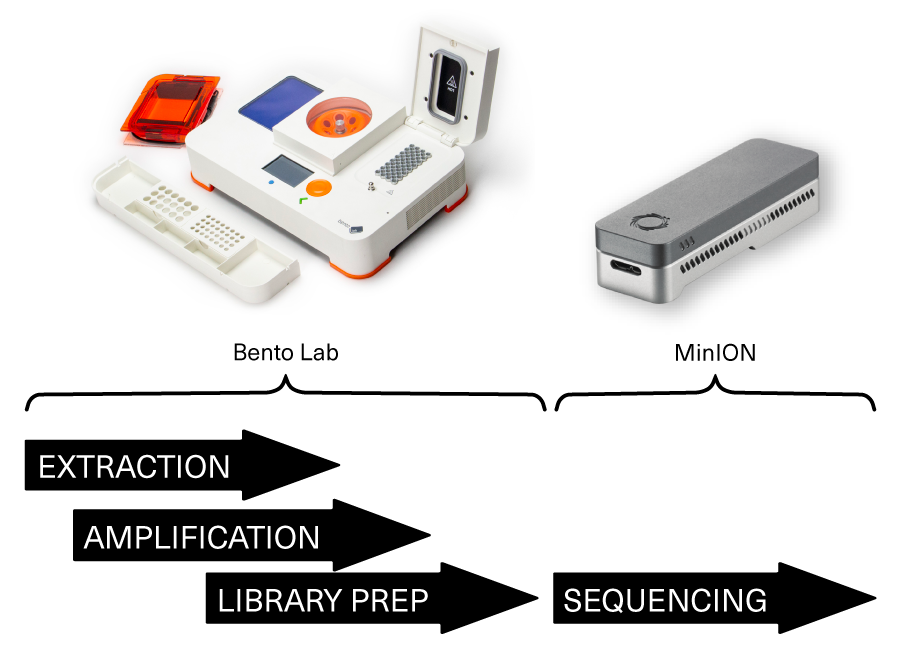
- All-in-one system: Streamlines DNA extraction, PCR, and gel electrophoresis for efficient nanopore sequencing preparation.
- Accessible and affordable: Simplifies workflows, reducing equipment needs while maintaining high-quality results.
Nanopore sequencing workflow and library prep
Nanopore sequencing platforms in combination with a mobile laboratory such as the Bento Lab offer a promising solution for improving research equality in countries fighting malaria.
Setting up Nanopore Sequencing with Bento Lab
Researchers around the world are using Bento Lab as a portable workstation for PCR and library preparation in MinION sequencing workflows.
Bento Lab is a portable all-in-one workstation for DNA extraction, PCR amplification, and gel electrophoresis, ensuring high-quality results for sequencing.
Whether you’re optimizing workflows, working in the field, or running a small lab, Bento Lab streamlines sample preparation, saving time and space. It’s the perfect tool to make nanopore sequencing more accessible and efficient.

Here, we present a protocol to genetically identify nematodes using 18S SSU rRNA sequencing using the MinION, a portable third generation sequencer, and proof that it is possible to perform all the molecular preparations on a fully portable molecular biology lab – the Bento Lab.
Learn more about using Bento Lab with nanopore sequencing.
Download whitepaperFeatured case studies

McCulley et al. (2025)
Bird Flu Genome in Antarctica
Researchers sequenced HPAI H5N1 from a seabird in Antarctica using portable RT-PCR and viral genomics tools in the field.
Tracking Bird Flu in AntarcticaMcCulley et al. (2025)

Soares et al. (2025)
Sequencing Biofilms On-Site
Whole-genome sequencing of SABs on a Roman monument reveals microbial diversity, biodegradation genes, and antimicrobial resistance.
Explore On-Site Biofilm Sequencing
Pillay et al. (2024)
Rapid Detection of Pathogens and Antimicrobial Resistance
In‐field nucleic acid isolation with Bento Lab for sequencing with ONT MinION to detect pathogens and antimicrobial resistance in meat, lake water, and wastewater sludge.
Rapid Detection of Pathogens and Antimicrobial Resistance
Dr Nabil Amor, University of Tunis El Manar
Decoding the Leopard Genome
Using portable genomics tools for conservation biology, Dr Amor has proven Arabian leopards exist, and he runs training for North Africa’s next-gen scientists.
Decoding the Leopard Genome
Salindeho et al. (2024)
C. burmanni Chloroplast Genome Study
Complete chloroplast genome of Cinnamomum burmanni re-sequenced and annotated, confirming IR regions and gene structure.
Complete chloroplast genome of Cinnamomum burmanni
Holzschuh et al. (2024)
Genomic surveillance of Plasmodium falciparum
Using a mobile nanopore sequencing lab for end-to-end genomic surveillance of Plasmodium falciparum: A feasibility study
Mobile nanopore sequencing lab: A feasibility study
Noah Bryan, Bayview Secondary School
Rapid Water Quality Testing
Noah Bryan, a high school student from Bayview Secondary School in Ontario, has developed a groundbreaking portable water testing kit under the supervision of Dr Lawrence Goodridge at the University of Guelph.
Rapid Water Testing with Portable Technology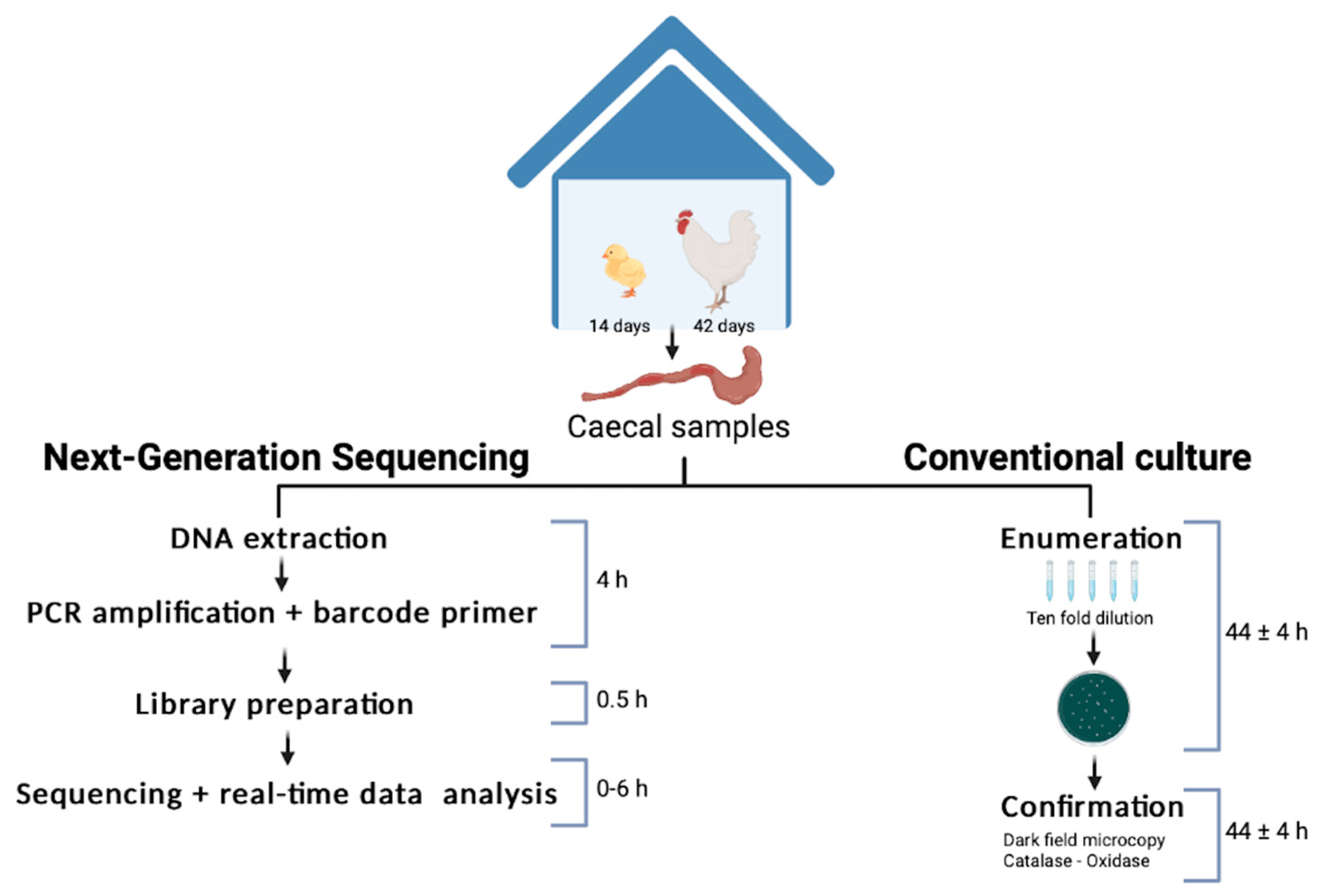
Marin et al. (2022)
Sequencing workflow for Campylobacter jejuni
Researchers used Bento Lab and MinION sequencing to develop a rapid on-site workflow for detecting Campylobacter in poultry. This method delivers results in under five hours — compared to the five-day standard — allowing real-time farm monitoring and improved food safety.
Sequencing workflow for Campylobacter jejuni
Chang et al. (2020)
Barcoding on a dive boat with Bento Lab and MinION
Can you DNA barcode specimens on a diving boat with Bento Lab and MinION, with all of your equipment packed into a backpack? And how many species could you sequence in one batch? A team of researchers from Singapore undertook the challenge to find out, and produced one of the first and largest reported examples of DNA barcoding with Bento Lab and MinION to date.
Barcoding on a dive boat with Bento Lab and MinIONChang et al. (2020) Takeaways from Mobile DNA Barcoding with BentoLab and MinION
Dr Cheryl L Ames, Tohoku University
Unlocking the Mysteries of the Wild Ocean
Dr Ames is is making it possible to predict the presence of specific marine species in different regions of the ocean. She uses Environmental DNA (eDNA for short), analysing traces of DNA left by species.
Unlocking the Mysteries of the Wild Ocean
Genome360, Genome Prairie
Agricultural Biosurveillance and Crop Protection
Genome Prairie’s mobile lab brings DNA analysis to Manitoba farms for on-site pest detection and empowering rural agricultural communities.
Agricultural Genomics with Genome360
Hirabayashi et al. (2021)
Tracking Antibiotic-Resistant Bacteria in the Field with Bento Lab and MinION
Could it be possible to track the spread of antibiotic-resistant bacteria with field equipment?
Researchers from Cambodia, Japan, and the UK used Bento Lab and Oxford Nanopore’s MinION to find out.
On-Site Genomic Epidemiological Analysis of Antimicrobial-Resistant Bacteria in Cambodia With Portable Laboratory Equipment

Dr Ineke Knot, NatureMetrics / University of Amsterdam
Portable Genomics at home and in the field
As part of her PhD, Ineke Knot used portable genomics to study parasites in great apes. Recently she joined biodiversity monitoring company NatureMetrics as a researcher, working from her home lab.
Ineke Knot InterviewDNA Barcoding of Nematodes Using the MinION
Protecting Great Apes with Portable Genomics

Dr Sophie Zaaijer, New York Genome Center
Rapid, on-site library preparation
Dr Sophie Zaaijer and her team have developed an inexpensive method to prepare ready-to-sequence libraries for MinION in 55 minutes, using Bento Lab.
Rapid re-identification of human samples using portable DNA sequencing
Gianpaolo Rando, BeerDecoded
Decoding the DNA of Beer for precision brewing
Gianpaolo Rando is on a mission to understand craft beer at molecular level. Inside a Swiss biohackerspace he prepares DNA libraries for sequencing 1,000 beer microbiomes with the Bento Lab.
Decoding Beer with Bento Lab
Recommended for nanopore sequencing applications
Bento Lab Pro
From DNA extraction, to PCR, to MinION library prep, using one portable workstation.
32 well thermocycler
Heat or incubate, lyse cells, amplify DNA via PCR or isothermal methods, ligate sequencing adaptors.
8k G centrifuge, 6 tube capacity
Sediment samples, spin down tubes, extract DNA using spin columns
Electrophoresis and transilluminator
Cast, load, and run agarose electrophoresis gels to visualise genomic DNA or amplicon DNA
